EMIF NEWSLETTER (JUNE 2017)
June 2017
Use the buttons below to jump to each article in this issue.
To Subscribe to the Newsletter, please complete the Enquiry Form.
| Article 1 OMOP |
Article 2 EMIF-AD |
Article 3 CASE 12 |
Article 4 NAFLD |
Article 5 EMIF-PLAT |
| JUNE 2017 |
01 |
|||
The Power of Harmonization:
|
||||

|
KEY POINTS | |||
 |
Data harmonization is the process of translating data into a common format and common vocabulary to enable collaborative research and large-scale analytics. |
|||
 |
The European Medical Information Framework (EMIF) has a core objective of building an information framework for consistent re-use of available patient-level data to support novel research. |
|||
 |
The Observational Health Data Sciences and Informatics (OHDSI) collaborative works toward the international implementation of the OMOP Common Data Model (CDM) and its standardized vocabularies.
|
|||
 |
EMIF is currently mapping European data sources to the OMOP CDM, and is extending OHDSI tools to enable the large-scale analytics that are key to EMIF’s core objectives.
|
|||
| CONTRIBUTORS | ||||
 |
MICHEL Van SpeybroeckDirector of Data Sciences at Janssen  |
|||
 |
PETER Rijnbeek, PhDAssistant Professor at Erasmus University Medical Center  |
|||
 |
JOHAN van der LeiAcademic Coordinator EMIF-PLAT, Professor at Erasmus University Medical Center  |
|||
Big Healthcare Data |
1A | |||
Every moment, countless healthcare data points are generated and stored in digital databases across Europe. Analyzing these datasets is a challenge, because data structure and content can vary greatly from one healthcare organization to the next. Data is collected for different purposes: clinical research, patient care, payer reimbursement, and more. This means that the content and level of detail varies considerably. Datasets are stored in different formats using different database management systems. Moreover, different codes are used for the same medical concepts across Europe, which creates additional challenges.
How can we overcome these overwhelming barriers to performing multi-database studies across Europe and beyond?
EMIF Rises to the Challenge |
1B | |||
The European Medical Information Framework (EMIF) recognized these challenges from day one. Standardization was initially done in the context of a specific research question on a limited set of variables. However, EMIF realized that this approach was difficult to maintain when answering research questions on a large scale. A substantial upfront standardization effort would be necessary for scalability. Additionally, studies needed to be performed in a distributive manner to comply with governance and privacy rules by keeping standardized datasets within their local infrastructures. The only shared analytical datasets needed to be generated using a standardized processing pipeline.
An Odyssey Toward Harmonization |
1C | |||
EMIF found the solution to its challenges by embracing the Observational Health Data Sciences and Informatics (OHDSI, pronounced “Odyssey”) model of translating all the data into a single common structure and language. This concept is known as “data harmonization” (also sometimes called “data standardization”).
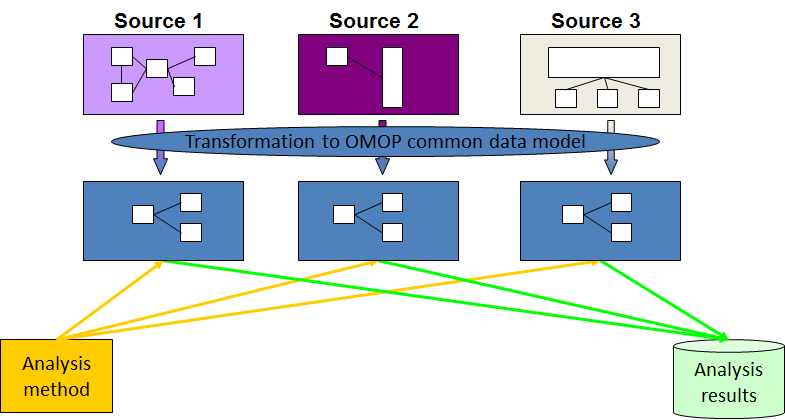
Diagram representing the process of data harmonization using the OMOP Common Data Model.
OHDSI grew out of the Observational Medical Outcomes Partnership (OMOP), a public-private partnership originally established in the US. The OMOP project developed the initial version of the OMOP Common Data Model, which can accommodate healthcare data from diverse sources in a consistent and standardized way. OHDSI’s mission is to improve health, by empowering a community to collaboratively generate the evidence that promotes better health decisions and better care. A key objective is to stimulate transparent and reproducible research for which the CDM is a prerequisite.
Today, OHDSI is an international collaborative comprised of academics, industry scientists, healthcare providers, and regulators who have come together to create and apply open-source data analytic solutions to a large network of health databases. It’s a non-funded entity, supported by the generosity and passion of individual members as well as in-kind contributions from organizations such as Columbia University and Johnson & Johnson. An inspiring spirit of global collaboration is what powers OHDSI.
EMIF and the Common Data Model |
1d | |||
EMIF is embracing OHDSI and the OMOP CDM for many reasons. The OMOP CDM is a well-adopted model covering over 600 million patient lives worldwide. All the code and tools are open source and publicly available, which stimulates collaborative tool improvement and transparency. Moreover, the currently available tools fit naturally with EMIF’s objective to facilitate different levels of data access.
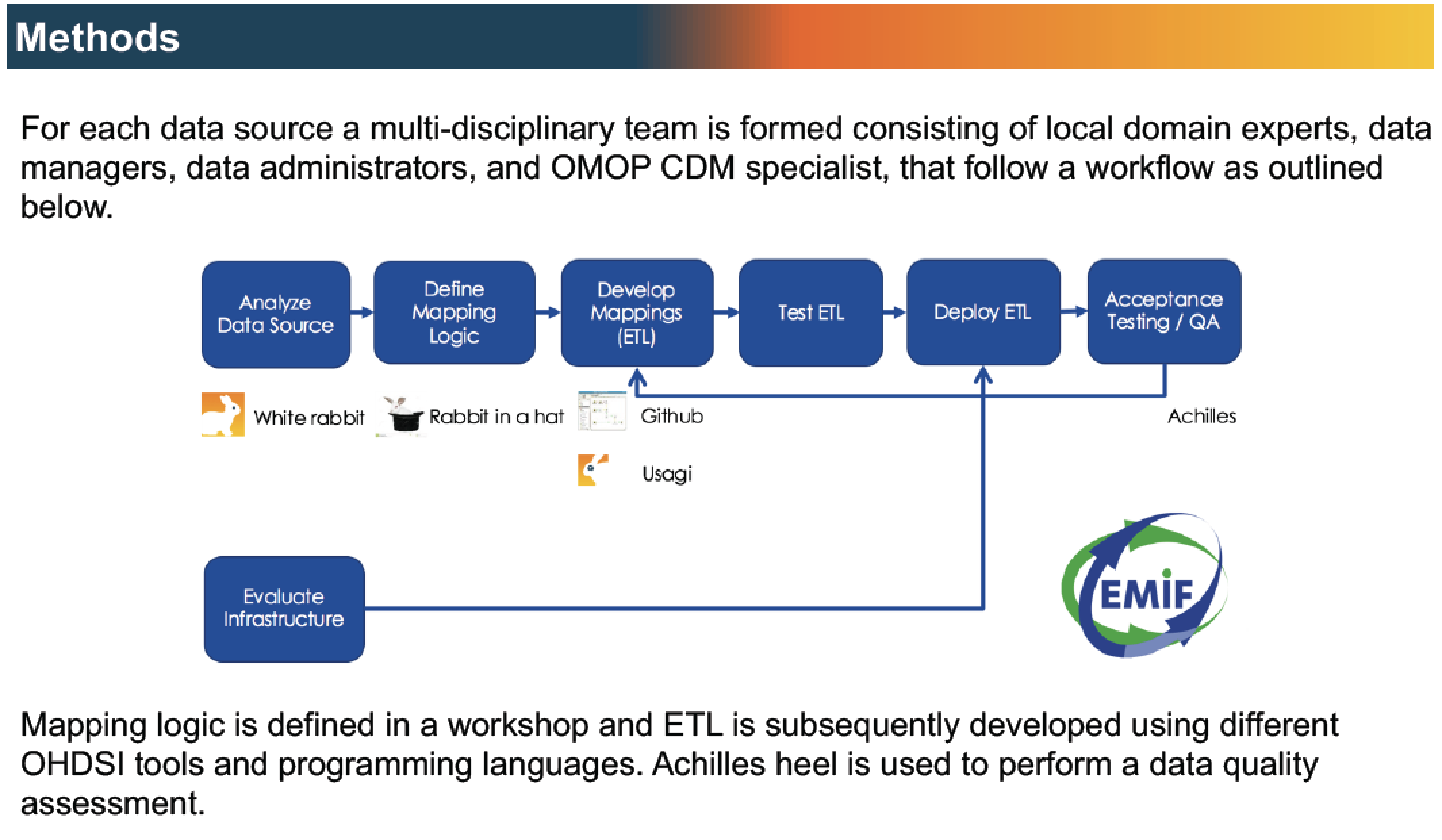
Workflow to map EMIF EHR data sources to the OMOP CDM, leveraging OHDSI tools and experience.
The CDM is most widely used in the US and Asia, and we are only beginning to scratch the surface of its use in Europe. We at EMIF are helping to prototype the CDM on the European continent. For example, we’re exploring whether there are levels of detail in some European Electronic Health Records (EHR) that would be lost if we translated data to the CDM. Our peers and fellow collaborators at OHDSI are constantly adjusting the CDM and its vocabularies to accommodate those types of insights.
The CDM is also evolving as healthcare evolves. As the OMOP Common Data Model expands to other continents, countries, and disease areas, the CDM expands thanks to the collaboration of OHDSI champions around the globe.
Calling All Data Custodians |
1F | |||
EMIF wants to actively support the adoption of the OMOP CDM across Europe. If you are a data custodian in Europe who wants to participate in this innovative data network, contact Michel Van Speybroeck, Peter Rijnbeek, or Johan van der Lei to learn more. While mapping your data does require a time commitment, EMIF is available to help. Additionally, there are resources available on the ODHSI website and training workshops offered during the free-to-attend OHDSI symposia.
Documents with best practices for converting data into the OMOP CDM are available on the OHDSI wiki page.
There are clear advantages to adopting the tools that ODHSI and EMIF are working on. Thanks to the multi-disciplinary expertise of the OHDSI community, the quality and functionality of the tools is constantly being improved. And the process is open source from start to finish, so no licenses are ever needed.
Perhaps most importantly, all data privacy concerns are respected by keeping your data local. You are implementing the CDM yourself. Even if we at EMIF provide support with the technical side of the process, your data is still kept locally. You’ll be able to maintain the privacy of your database while contributing to a worldwide effort to harmonize data. Your decision to map your database to the CDM means that you are supporting the progress of reproducible scientific research on a local, national, and international level.
Next Steps at EMIF |
1G | |||
EMIF is actively mapping nine European data sources to the OMOP CDM and OHDSI vocabularies: Denmark (AUH), Italy (ARS, IMS HEALTH LPD, PEDIANET), Spain (IMASIS, SIDIAP), UK (THIN), Estonia (EGCUT), and the Netherlands (IPCI, PHARMO). EMIF will produce a scientific paper this year that evaluates the mapping process of these European datasets and formulates suggestions for improving the CDM and its vocabularies. Furthermore, EMIF will continue the effort of extending and refining the OHDSI tools to meet its objectives.
If we want data analysis at an unprecedented scale, along with fully transparent and reproducible research pipelines, we need to collectively harmonize our data structures and terminology. The OMOP CDM is clearly becoming the de facto standard for electronic health data across the world. EMIF recognizes this exciting new reality, from which all stakeholders will benefit: researchers, organizations, and most importantly patients—who are ultimately at the center of this global effort.
If you are a researcher and want to explore more, please visit the EMIF Catalogue. If you are a data custodian interested in learning more about the OMOP-CDM for your data source, contact Michel Van Speybroeck, Peter Rijnbeek, or Johan van der Lei.
| JUNE 2017 |
02 |
|||
With Knowledge Comes Responsibility: Harmonizing Cohort Data for EMIF-AD |
||||

|
KEY POINTS | |||
 |
The European Medical Information Framework (EMIF) Catalogue allows users to quickly complete new research by identifying cohorts via the EMIF Alzheimer’s disease (EMIF-AD) community. |
|||
 |
Analyses of large pooled datasets are made possible by the EMIF-Platform technical team who developed the idea of using knowledge objects for the purpose of data harmonization. |
|||
 |
The EMIF-AD team invites data custodians and researchers to provide metadata about their cohort and share more data and research questions to help crack the dementia code. |
|||
| CONTRIBUTORS | ||||
 |
RUDI VerbeeckSenior Manager at Janssen R&D IT  |
|||
 |
ISABELLE BosNeuropsychologist and PhD Candidate at Maastricht University  |
|||
 |
LUIZA Gabriel-ZeltzerPostdoctoral Researcher at Janssen R&D IT  |
|||
 |
STEPHANIE VosNeuropsychologist and Postdoctoral Researcher at Maastricht University  |
|||
 |
PIETER Jelle VisserAssociate Professor, Maastricht University, VU University Medical Center, Project Lead of the AD topic  |
|||
The Biomarker Discovery Study |
2A | |||
Within EMIF-AD, projects are divided into work packages (WPs). WP 1 focuses on the “definition of study population data requirements and data collection.” Members of the EMIF-AD WP 1 team (Pieter Jelle Visser, Stephanie Vos, and Isabelle Bos) consulted the EMIF Catalogue AD community, an online community of existing cohorts maintained by EMIF‐AD. They identified 11 suitable cohorts with participants who had the characteristics the researchers were looking for in terms of age, sex, cognitive status, amyloid biomarkers in cerebrospinal fluid (CSF) or on positron imaging tomography (PET), and availability of MRI scans and fluid samples in plasma or CSF.
The EMIF-AD WP 1 team contacted principal investigators of these cohorts and all of them were interested to participate. Contracts were set-up to allow data-sharing and biomarker analysis. Data source owners harmonized—or translated—their local variables to a global set of variables so that the data could be used in the same study. In this way, a large cohort was assembled quickly and inexpensively.
The final Biomarker Discovery Study included 1,221 subjects. The data combined proteomics, genomics, metabolomics, and imaging data in a single analysis. This broad spectrum analysis in a large cohort of subjects at various cognitive stages will provide new insights into underlying mechanisms and facilitate the discovery of new diagnostic and prognostic markers for AD. This study is coordinated by Simon Lovestone, Johannes Streffer, and Pieter Jelle Visser in close collaboration with proteomics, metabolomics, transcriptomics, genomics, bioinformatics and imaging experts in EMIF-AD.
Translating the Language of Science |
2B | |||
The Biomarker Discovery Study and other analyses of large cohorts were made possible by a collaboration between EMIF-AD researchers and the EMIF-Platform technical team who developed the idea of using knowledge objects for the purpose of data harmonization. A knowledge object is a variable, plus all the information that you need in order to correctly interpret what that variable means. For instance, “sex” is a knowledge object included in the Biomarker Discovery Study, and “male” and “female” are common values for that knowledge object.
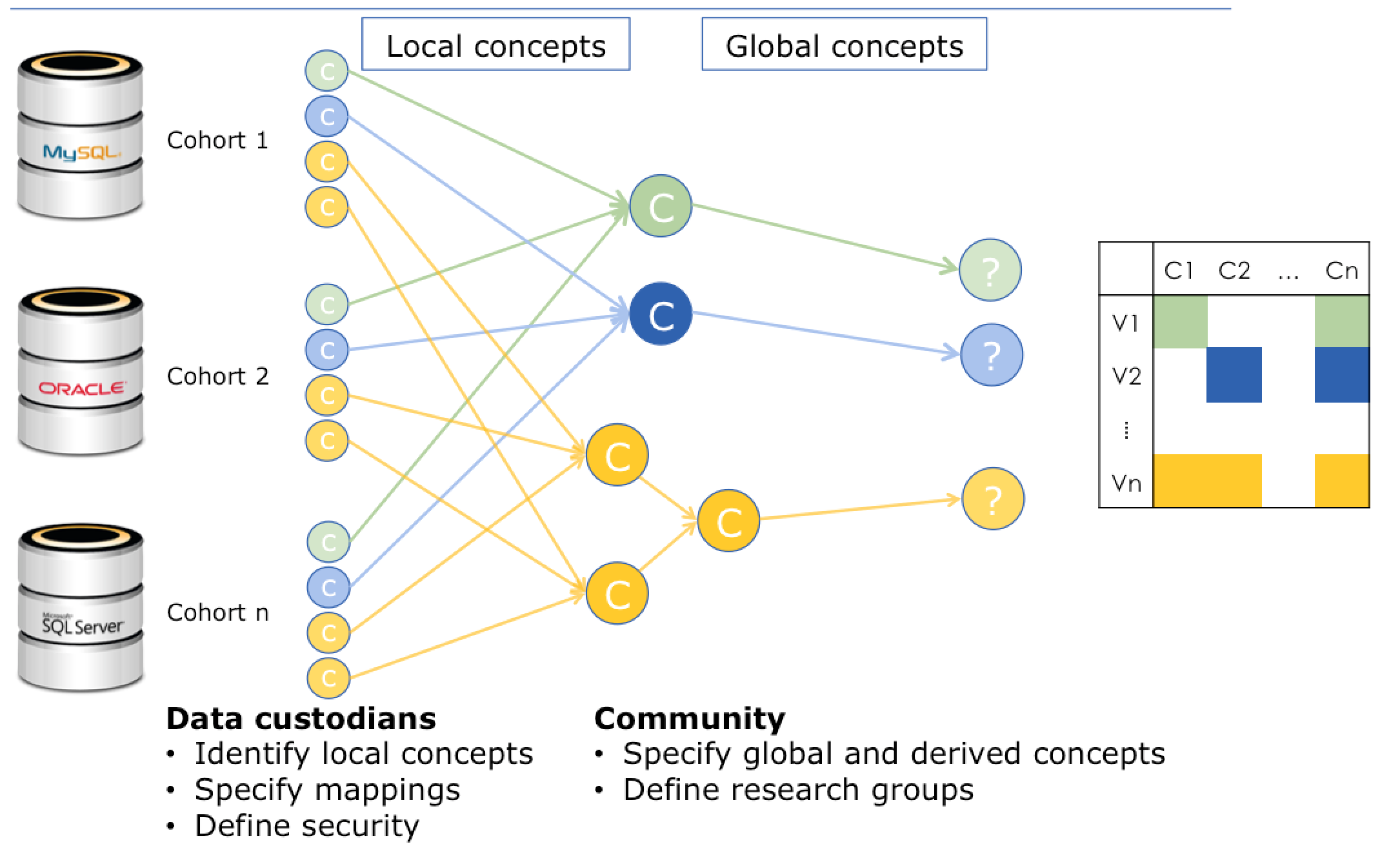
Diagram envisioning the process of data harmonization, mapping local knowledge objects to global knowledge objects. Data sources appear on the left, while harmonized data appears on the right.
When the EMIF-AD researchers gathered the data from 11 different studies to assemble the EMIF-AD WP3 1000 cohort, they were in different languages and none of them used exactly the same standards for recording variables. If they had tried to access the original data from the data sources, the variable “sex” might have appeared as “genero” or “geschlecht.” More obscure variables may have been difficult to interpret or impossible to translate to similar datasets without a common reference point.
That’s why Rudi Verbeeck, Senior Manager at Janssen R&D IT and EMIF contributor, believes cohort data custodians play a crucial role in the quest to aggregate data. Only a data custodian knows what the variables in their data source truly represent. Thanks to the EMIF-Platform team, custodians now have the power to define their local variables so their data can be translated to a standard set of global variables.
Knowledge and Responsibility |
2C | |||
“If you have the knowledge, you have the responsibility,” explains Rudi. Data custodians have knowledge about their data, so they are responsible for defining their local variables. Researchers know what points of data support their research, so they are responsible for identifying the variables they want to access. But who brings together the knowledge of the data custodians with the knowledge of the researchers so that data can be harmonized and large cohorts can be analyzed?
Isabelle Bos and Stephanie Vos, neuropsychologists and researchers at Maastricht University, are members of the EMIF-AD WP 1 team. They’ve chosen to take on this responsibility of helping data custodians translate their local knowledge objects to global knowledge objects. If data custodians don’t have the technical skills to map the data themselves, the data custodian specifies how the mapping should be done, then the EMIF-AD WP 1 team creates the code that needs to be executed. There are currently 14 cohorts on the data platform tranSMART from across Europe (Netherlands, UK, Belgium, Spain, Italy, Sweden, France, Greece). The total number of subjects across all 14 cohorts is 3,423, and this number keeps growing.
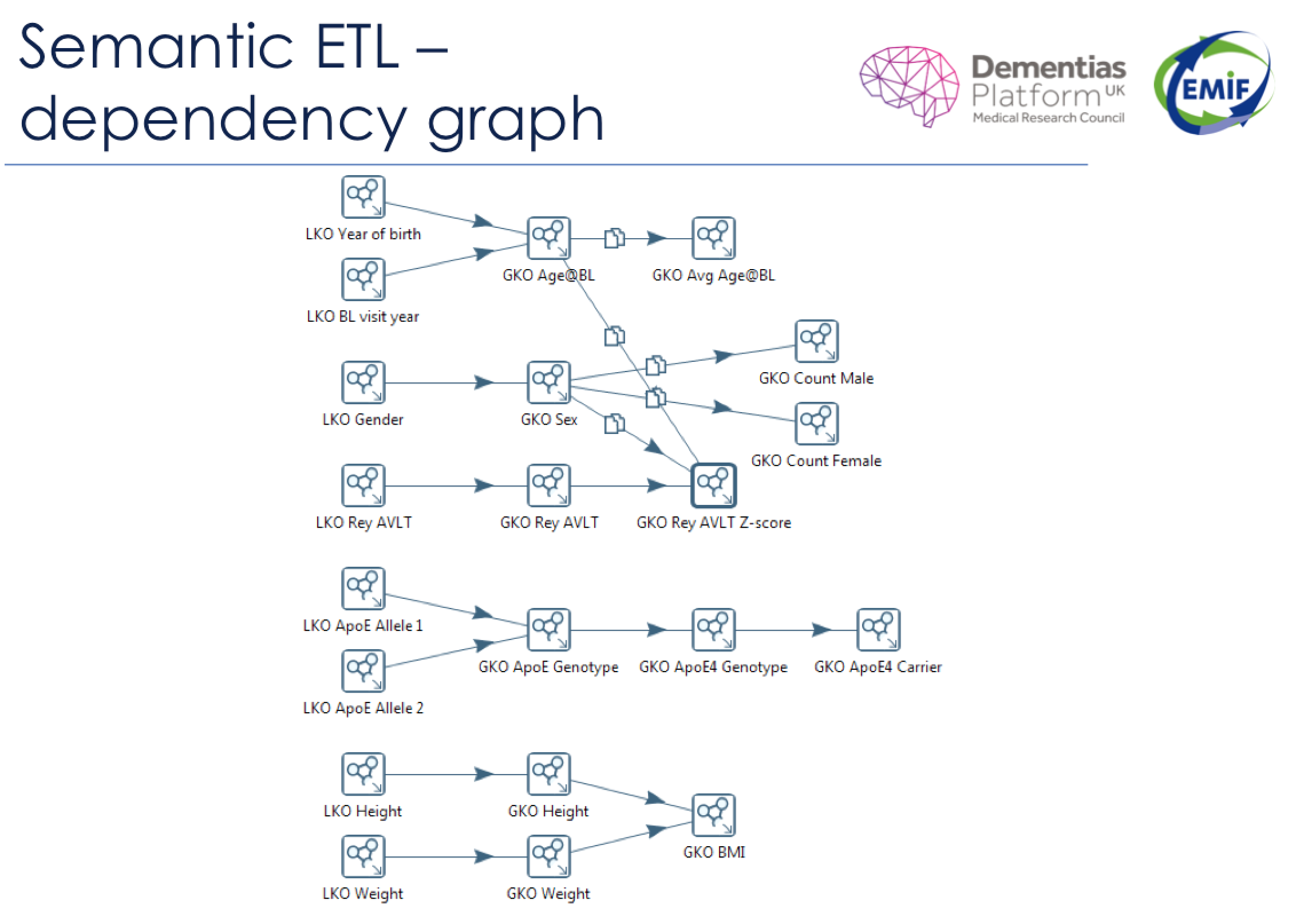
Local knowledge objects (LKO) appear on the left side of this semantic dependency graph. The local knowledge objects are translated to global knowledge objects (GKO) using snippets of code (represented by the arrows). Sometimes, there is a one-to-one correlation between a local knowledge object and a global knowledge object. Other times, multiple local knowledge objects feed into a single global knowledge object through a series of standardizing equations.
Building a Library |
2D | |||
Over time, EMIF-Platform will continue building its library of reusable knowledge objects and the snippets of code that connect them. If a researcher makes a request in the EMIF Catalogue AD community for variables that haven’t been used before, those knowledge objects will need to be mapped by the data custodians with support from the EMIF-AD WP 1 team.
Since every research question adds to the library, the probability of being able to reuse a knowledge object or snippet of code increases with every research question that comes along.
Our Common Goal |
2E | |||
In order to tackle some of the biggest health challenges of our time, researchers must be able to conduct efficient and cost-effective data analyses on a large scale. For that, data harmonization is a necessity. With a cohort of 100 subjects, you can’t answer complex research questions—but increase that cohort to 1,000, and you may uncover the kind of exciting results that the EMIF-AD group and their collaborators are seeing in the Biomarker Discovery Study.
Before we can harmonize data, it needs to be shared. If you own cohort-derived data—especially if it includes biomarkers—and would like to collaborate, please consider working with the EMIF-Platform team to harmonize your data using knowledge objects. Harmonized cohort data could lead to other breakthroughs in the fight against Alzheimer’s.
If you are a researcher, explore the data sources already available through the EMIF Catalogue AD and EHR communities, which were made publicly available to bona fide researchers outside of EMIF in January 2017. Every day, new people from around the world join the 309 users who have accessed the Data Catalogue as they work toward their research goals. If you could assemble huge cohorts quickly and inexpensively, what kinds of studies would you create? What discoveries might you spearhead? EMIF wants to know and wants to help you make them happen.
| JUNE 2017 |
03 |
|||
Introducing Use Case 12:
|
||||

|
KEY POINTS | |||
 |
By analyzing electronic health records (EHR) data through the European Medical Information Framework (EMIF) platform, we can find out more about the link between inflammation and dementia. |
|||
 |
Use Case 12 is utilizing EMIF’s visible, accessible, harmonized datasets to complete ground-breaking research about whether inflammatory diseases or the drugs used to treat them change the risk of dementia in later life. |
|||
 |
Later in 2017, look for published information that will likely assist in the development of therapies and treatments for dementia. |
|||
| CONTRIBUTORS | ||||
 |
DANIELLE NEWBYPost-doctoral researcher at University of Oxford, Data analyst of Use Case 12  |
|||
 |
ALEJO NEVADO-HOLGADOSenior post-doctoral researcher at University of Oxford, Lead of Use Case 12 project  |
|||
 |
SIMON LOVEStoneProfessor of Translational Neuroscience at University of Oxford, Overall joint program lead for EMIF  |
|||

Challenges and Solutions |
3A | |||
The motivated team at Oxford was curious about the relationship between inflammatory diseases, anti-inflammatory drugs, and dementia. They saw an exiting opportunity to work from large, real-world, routine care datasets, but realized such studies are challenging to perform. For one thing, large datasets are needed for analysis. For another, research study cohorts can suffer from selection and are not always fully representative. “If large datasets are accessible,” Danielle emphasizes, “then another challenge presents itself: it’s tough to get consistent datasets due to the different structures of electronic health records (HER) in different countries.”
EMIF provided an opportunity to overcome these problems by making datasets visible, accessible, and harmonized. Through Use Case 12, the Oxford team is now examining the relationship between diseases of inflammation and the drugs used to treat inflammatory disorders in multiple datasets. Through this work, they’re revealing stronger evidence of the relationship between dementia and inflammatory exposures. The team’s ground-breaking research sets an example for others. Other researchers may be excited to learn that the questions like those posed within Use Case 12 can be analyzed and answered efficiently by working with EMIF.
From Start to Finish |
3B | |||
To start Use Case 12, the team needed a good question that could potentially be answered using EHR data. With a hypothesis relating to inflammation and dementia, their protocol underwent ethical and peer review by experts in the field.
The next step was to contact the data custodians responsible for some of the datasets in the EMIF Catalogue; those who were able to participate joined Use Case 12. The team then asked the data custodians to extract the variables they needed from anonymized patient level records from their databases and place them on a secure remote research environment. Daniel Prieto-Alhambra, scientific coordinator for the SIDIAP database, was enthusiastic about contributing the SIDIAP database for Use Case 12, stating that “the inclusion of the SIDIAP dataset into Use Case 12 offers a great opportunity to utilize the wealth of information from this database to help understand such an important topic.”
After quality control checks for each of the datasets, local database experts started their work. For each dataset, they ran a software tool which aggregated the files for each dataset, removed any outliers, and selected the sample population based on the criteria specified in the original protocol. Thanks to a seamless collaboration with the EMIF-Platform team, the data for each dataset at this stage was then ready for statistical analysis.
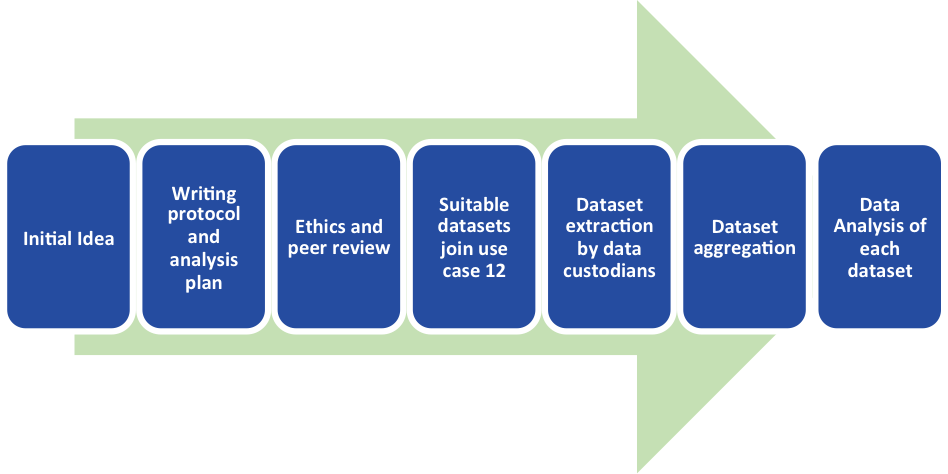
The Workflow of Use Case 12
Where Are We Now? |
3C | |||
Use Case 12 is on the cusp of the most exciting phase: data analysis! Nearly all the datasets have been extracted and are almost ready to be analyzed. The team built a clever step into their process; they are analyzing one dataset first, so they can run preliminary checks and help troubleshoot the analysis of the rest of the datasets.
Alejo confirms that participating in Use Case 12 has been “an excellent learning experience for our team” when it comes to the opportunities and challenges of analysis across multiple real-world datasets. “We have been fortunate to have good help and expertise to guide us and move the work forward successfully,” Danielle reports. Later in 2017, look for a publication of the findings from all of the datasets in Use Case 12. The team believes the results could lead to meaningful advancements in the treatment of Alzheimer’s disease and dementia.
Take Action |
3D | |||
The EMIF use cases are a perfect example of successful collaborations and data sharing between and among researchers and data custodians. If you have an idea that you think could be answered using EHR data, then the progress of Use Case 12 should give you confidence that a robust analysis is possible. If you would like to learn more and get involved in the continuing progress of Use Case 12, contact Danielle or Alejo to collaborate or network. They welcome ideas, help, and expertise from anyone within EMIF who is interested in being involved with dementia research. Danielle, speaking on behalf of the Use Case 12 team, says: “We want to collaborate with you and share our knowledge to make even greater progress toward our ultimate goal: the development of new therapies and treatments for dementia.”
For further reading about determining the molecular pathways underlying the protective effect of non-steroidal anti-inflammatory drugs for Alzheimer’s Disease, please see Alejo and Simon’s October 2016 publication.
| JUNE 2017 |
04 |
|||
Precompetitive Research by a Multi-Disciplinary Team: The Investigation of Non-Alcoholic Fatty Liver Disease as Recorded in Primary Care Databases |
||||
|
KEY POINTS | |||
 |
The European Medical Information Framework (EMIF) enables collaboration between partners from the pharmaceutical industry and academia. |
|||
 |
EMIF collaborators conduct precompetitive research in multiple European databases in a unified environment. |
|||
 |
EMIF enables the pharmaceutical industry to have unprecedented access to routinely recorded European healthcare data while respecting appropriate security and ethics provisions. |
|||
 |
A cross-company team discovered heterogeneity in the prevalence of recorded non-alcoholic fatty liver disease in European databases, as well as a low detection rate in primary care. |
|||
| CONTRIBUTORS | ||||
 |
MYRIAM ALEXANDERClinical data scientist and analyst at GlaxoSmithKline  |
|||
 |
NAVEED SATTARProfessor of Metabolic Medicine at the University of Glasgow  |
|||
 |
DAWN Genetics Director at GlaxoSmithKline
|
|||
 |
KATRINA loomisHead of Cardiovascular and Metabolic Disease Human Genetics at Pfizer  |
|||
 |
WILLIAM alazawiSenior lecturer and Consultant in Hepatology at the Blizard Institute, Barts and The London School of Medicine  |
|||
 |
JOLYON fairburn-beechData analyst at GlaxoSmithKline  |
|||
Our Motivating Questions |
4A | |||
Prior to collaborating, each of us was motivated to answer key questions about the epidemiology of non-alcoholic fatty liver disease (NAFLD), such as:
- What is the prevalence and incidence of the disease in primary care data?
- Is NAFLD associated with an increased risk of cardiovascular diseases after accounting for risk factors that may potentially confound this association?
- What excess risks do NAFLD patients experience when it comes to developing liver complications, including cirrhosis and hepatocellular carcinoma?
These questions remained difficult to answer without access to a large sample of patients with extensive medical data. Fortunately, we had the opportunity to work within EMIF.
Unprecedented Access |
4B | |||
Via EMIF, we have access to unprecedented large-scale databases collected routinely by primary care systems of four European countries:
- The UK: The Health Improvement Network (THIN)
- The Netherlands: Interdisciplinary Processing of Clinical Information (IPCI)
- The Catalonia region of Spain: Information System for the Development of Primary Care Research (SIDIAP)
- Italy: Health Search – IMS HEALTH Longitudinal Patient Database (Health Search IMS HEALTH LPD)
We identified suitable databases using the EMIF Catalogue EHR community, then used the support of EMIF to contact data custodian project partners.
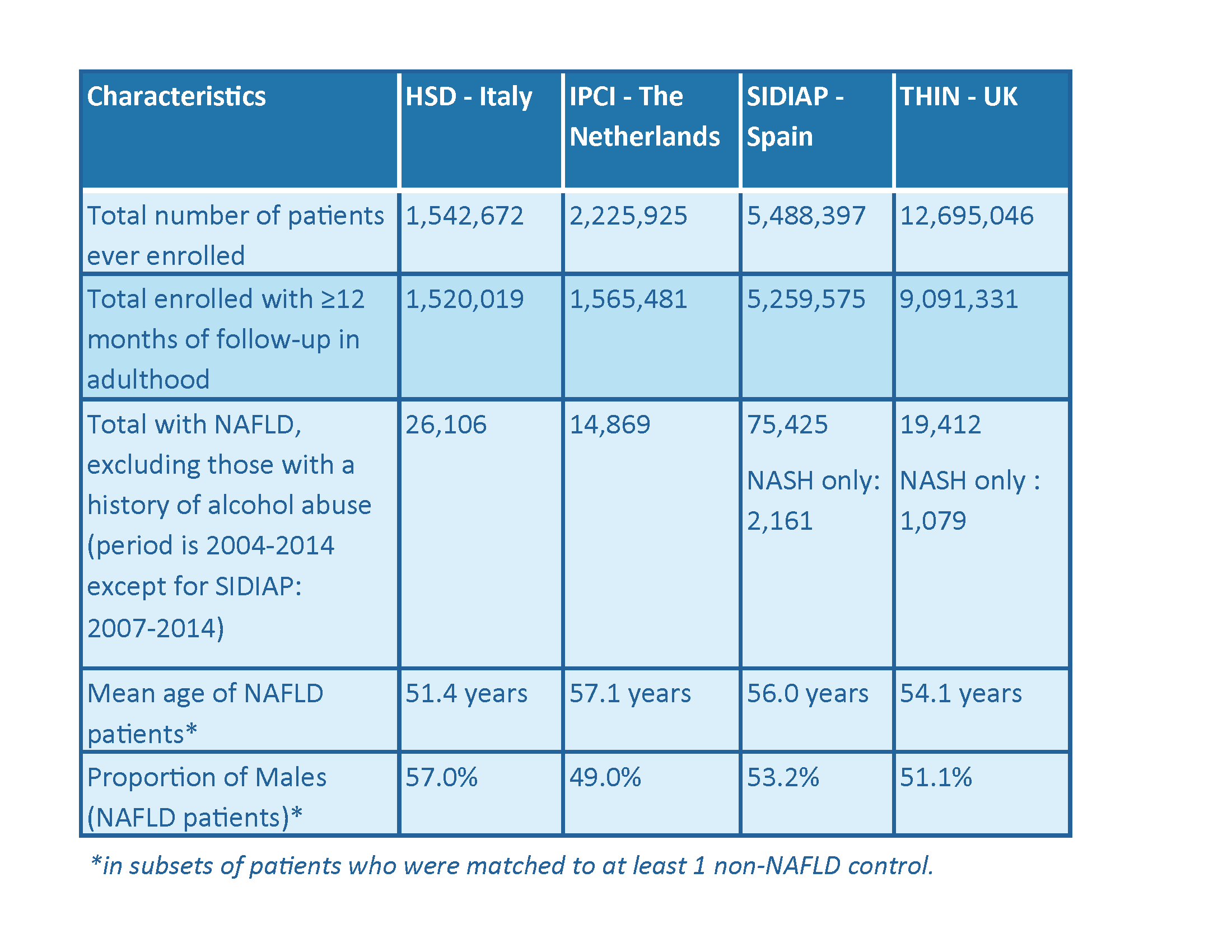 Attrition and demographic characteristics of patients with NAFLD in four European databases.
Attrition and demographic characteristics of patients with NAFLD in four European databases.
Harmonizing the Data |
4C | |||
For Use Case 10, our team developed a protocol and submitted it to the ethical review bodies of each database; upon approval, a single finalized protocol was circulated to all parties. The four databases used different terminology systems to code clinical diagnoses, so a process of data harmonization was needed to enable the team to create comparable lists. The protocol was then translated into a series of data management and statistical analysis steps, after which data was locally extracted by each data custodian and centrally analyzed in a remote server.
A New Level of Trust |
4D | |||
Thanks to EMIF, the pharma companies involved in the project have been able to access—for the first time—an anonymized subset of the raw data of these databases. From there, the companies have been able to perform the analyses, plumbing the depths of the various databases. This has enabled all parties to experience a new level of trust in the efficient reuse of routine data for the benefit of bona fide research.
First Results |
4E | |||
The analysis by a data analyst at GlaxoSmithKline (GSK) is ongoing. A late-breaking abstract was presented at the European Association for the Study of the Liver (EASL) 2017 conference on incidence and prevalence of NAFLD. Data were available for 17.4 million adults with more than 1 year of follow-up from registration. Over 135,000 individuals had a diagnosis of NAFLD or non-alcoholic steatohepatitis (NASH), amounting to a pooled prevalence of 1.7% as of January 2014. Although these are relatively low compared to prevalence estimates in the literature of 20 – 30% of NAFLD in the general population, patients’ characteristics were in line with the literature.
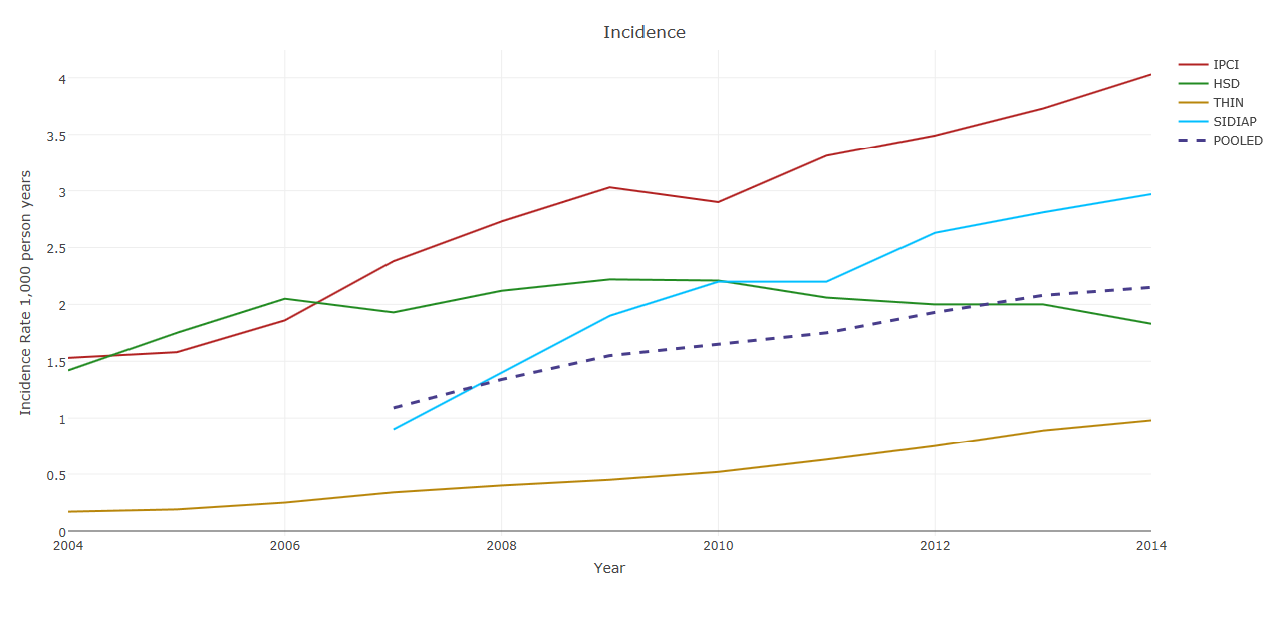
Incidence of recorded NAFLD (per 1,000 person-years) in 2004–2014 in the four countries, and pooled across countries.
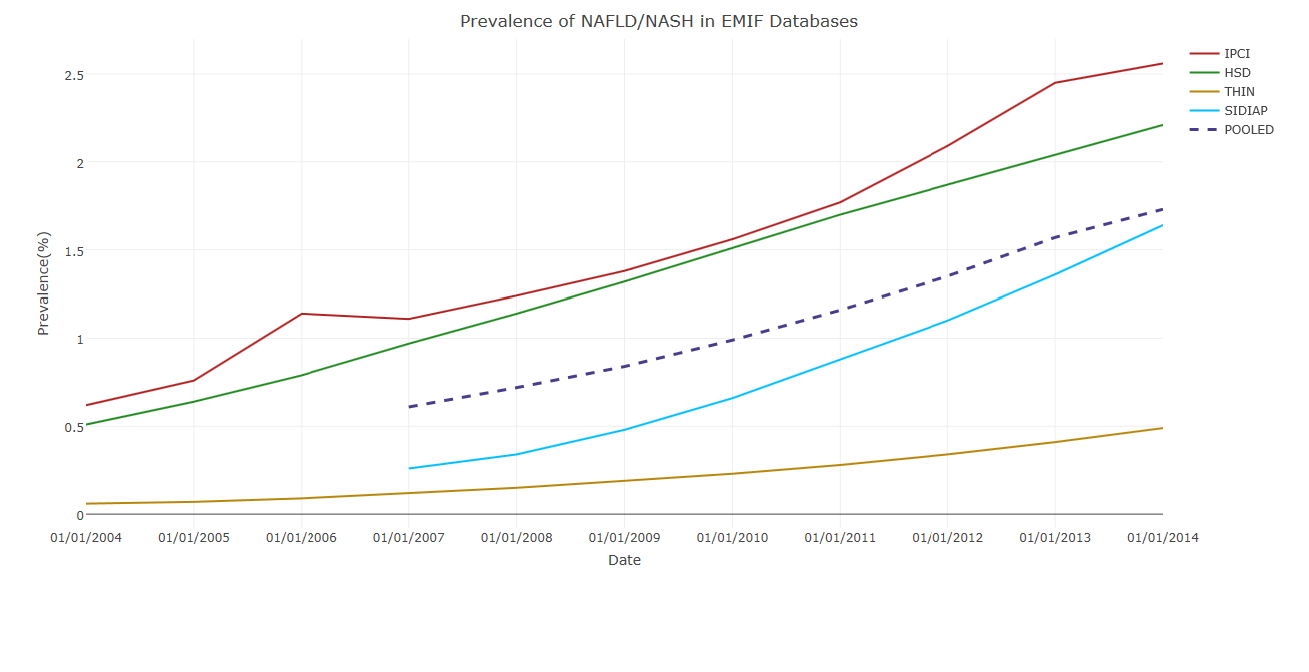
Point-Prevalence of recorded NAFLD (per 100 persons) in 2004–2014 in the four countries, and pooled across countries.
Next Steps |
4F | |||
This sample of NAFLD patients represents the largest NAFLD cohort ever assembled. It will enable further research on NAFLD, which is of interest to the pharmaceutical industry and the field of medicine. Our team is especially excited about potential research that may answer the motivating questions we mentioned above. We want to know if NAFLD is associated with an increased risk of cardiovascular diseases after accounting for risk factors that may potentially confound this association, and we also want to know what excess risks NAFLD patients experience when it comes to developing liver complications.
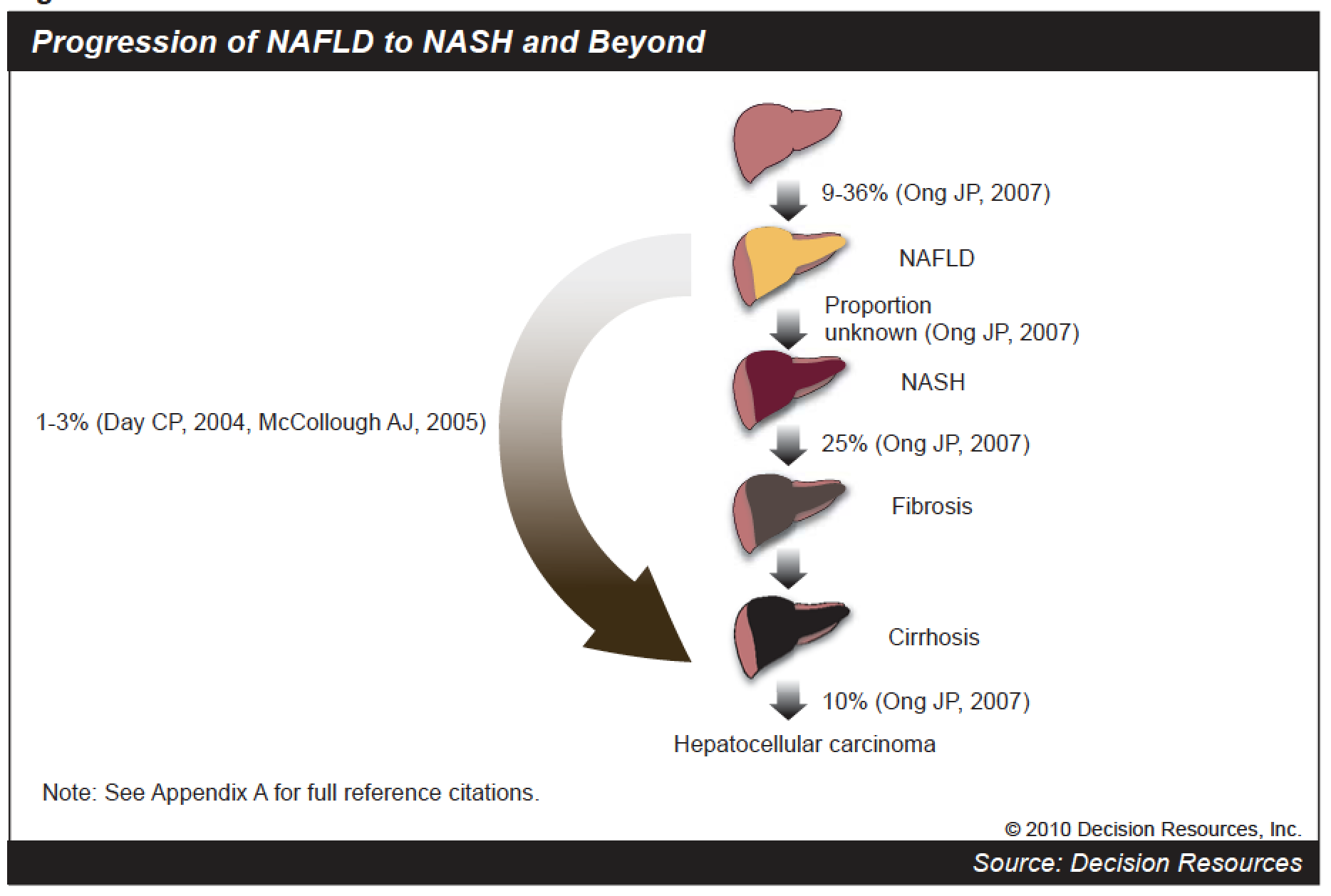
Progression of NAFLD to NASH and beyond (source: Decision Resources).
If you’re in the field of metabolics and want to learn more about this study and its findings, contact Myriam.
| JUNE 2017 |
05 |
|||
A Milestone Meeting: EMIF-Platform’s 9th Consortium Meeting |
||||

|
KEY POINTS | |||
 |
The exciting 9th Consortium Meeting for the European Medical Information Framework (EMIF) Platform took place in Barcelona, May 22–23, 2017. |
|||
 |
Important milestones were discussed, including the technology platform, data harmonization, sustainability, research use cases, expedited research efficiency, and publication strategy. |
|||
 |
EMIF is making significant contributions to the field of real world data for health research, but more work remains to be done in the final year of this Innovative Medicines Initiative (IMI) project. |
|||
| CONTRIBUTORS | ||||
 |
NIGEL HUGHESEFPIA Coordinator EMIF-PLAT, Janssen Pharma R&D  |
|||
Over the past four years, we at EMIF have made considerable progress. We have developed an integrated platform to facilitate access to diverse medical and research data sources across Europe. That sustainable framework now supports the identification, assessment, and use or reuse of health data for medical research.
To guide the development of that information framework, IMI originally selected two therapeutic areas that are critically important at this juncture in history: Alzheimer’s disease (EMIF-AD) and metabolic diseases (EMIF-Metabolic). These pressing research topics provided the EMIF-Platform with clear opportunities to support researchers with technological tools and governance processes. Thanks to the EMIF-Platform team, the researchers in EMIF-AD and EMIF-Metabolic are now working to address some of the world’s biggest health challenges.
A Meeting of the Minds |
5A | |||
Since EMIF-AD and EMIF-Metabolic leverage the framework created by the EMIF-Platform, one could say that the EMIF-Platform is the bedrock of EMIF. That’s why approximately 50 EMIF colleagues met in Barcelona for the 9th Consortium Meeting of the EMIF-Platform. On May 22 and 23, 2017, the EMIF-Platform team shared milestones, recognized progress, and discussed the work on the horizon for the rest of the year. Six major themes emerged.
Theme 1: Technology Platform |
5B | |||
For the first time in the four-year history of EMIF, an integrated platform was presented in outline form (see figure 1). While we will be evaluating this integrated platform for the rest of the year, its visualization is a major achievement for EMIF. This integrated platform allows researchers to access the EMIF Catalogue, specify parameters for the data that fits their research questions, and analyze that data in a secure private remote research environment (PRRE). This technological development means that researchers will be able to analyze large datasets while still respecting data privacy.
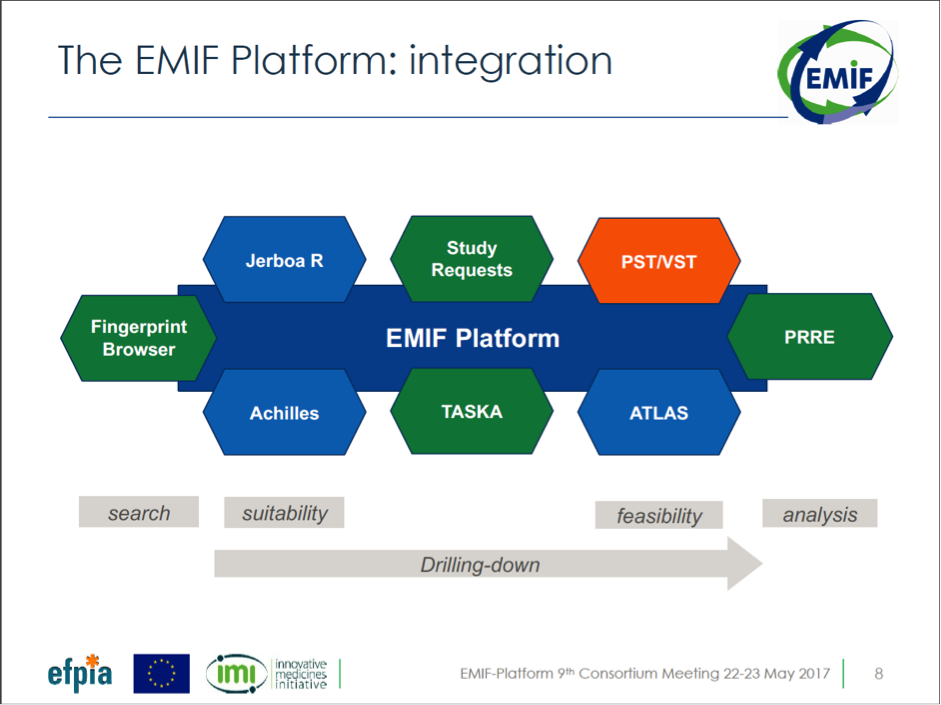
Figure 1: Integrated EMIF Platform
Theme 2: Data Harmonization |
5C | |||
The EMIF-Platform has helped the European continent advance towards data harmonization. We continue to map population data sources to the OMOP Common Data Model (CDM). The EMIF-Platform team is working hard to make sure that our data custodian colleagues are comfortable with the mapping conversion process. The EMIF-Platform team is also offering support when data custodians use the Observation Health Data Sciences and Informatics (OHDSI) tool set for mapping data to the CDM.
Simultaneously, we’re making progress assembling cohort data for EMIF-AD. Knowledge objects—variables that translate data from local to global vocabularies—allow us to add more AD cohorts to the EMIF Catalogue AD community. The knowledge objects team similarly supports data owners and researchers to add and access relevant cohort data.
Theme 3: Sustainability |
5D | |||
EMIF’s goal is to create a sustainable technology and governance framework for the identification, assessment, and use or reuse of health data for medical research in Europe. We’ve spent the past four years developing the technology and governance framework. Now, we’re increasing our focus on sustainability. While we’ve made progress toward developing potential sustainability models for the population and cohort tracks, much more work is needed before the end of this year. We need to have a clear plan to implement during EMIF’s proposed extension in the first half of 2018..
Theme 4: Research Use Cases |
5E | |||
During the 9th EMIF-Platform Consortium Meeting, colleagues shared the progress of many use cases (see figure 2), which will come to fruition later this year. Invaluable insights from these use cases will help EMIF better understand the research process, as well as potential challenges researchers and data custodians face. Stay tuned as use cases—like Use Case 10 and Use Case 12—are documented and shared.

Figure 2: EMIF-Platform Research Use Cases currently running
Theme 5: Expediting Research Efficiency |
5F | |||
Deeper insights into the research process—especially when it comes to bottlenecks and potential solutions—will enable EMIF to manage the research more efficiently. Insights from current use cases and proposed studies will allow us to pressure test operational feasibility, helping us evaluate the EMIF-Platform architecture from a scientific and research perspective.
Theme 6: Publication Strategy |
5G | |||
The EMIF-Platform consortium also focused on prioritizing publications for the remainder of the project. Within the remainder of the IMI program, we plan to promote external publications about EMIF’s technological, methodological, and scientific activities. We need to ensure the research community at large becomes familiar with EMIF and our developments, so we can invite interested parties to join us.
Progress and Future Work |
5H | |||
While discussions at the 9th EMIF-Platform consortium revealed that we have work to do in the limited time that remains, we also recognize that EMIF has made significant contributions to the field of real world data for health research. Our work over the past four years has proven how critical it is to have a sustainable, integrated platform for diverse data source collaborations, based on a harmonized approach to a federated network model.
All partners and collaborators can be rightfully proud of our achievements. Now we can focus our energy on all that needs to be done in the rest of 2017. We must create an operational model to sustain European medical research via real world data in 2018 and beyond.
If you would like to support the future of EMIF, please contact Nigel Hughes.


