EMIF-AD
EMIF-AD Project Background
With multiple clinical trials in dementia failing over the past years, attention has recently shifted to subjects with Alzheimer’s disease (AD) who have not reached the stage of dementia yet (i.e., ‘pre-dementia’). Since these subjects have limited brain damage and are suggested to be more likely responsive to treatment than subjects with full-blown dementia, they are an important target group for future treatment studies.
EMIF-AD Project Objectives
 Trial design in pre-dementia AD, however, is challenging because subjects with pre-dementia AD are difficult to identify and limited information is available on their outcome. The lack of reliable diagnostic and prognostic markers for pre-dementia AD can be explained by the availability of only small-scale ongoing biomarker studies and longitudinal cohorts including these subjects. EMIF has linked this information and unlocked the true potential of these studies.
Trial design in pre-dementia AD, however, is challenging because subjects with pre-dementia AD are difficult to identify and limited information is available on their outcome. The lack of reliable diagnostic and prognostic markers for pre-dementia AD can be explained by the availability of only small-scale ongoing biomarker studies and longitudinal cohorts including these subjects. EMIF has linked this information and unlocked the true potential of these studies.
By connecting relevant cohort studies across Europe, EMIF-AD has set up a pan-European platform for large-scale research on biomarkers and risk factors for neurodegenerative disorders. The biomarker discovery activities in EMIF-AD were driven by an extreme phenotype approach, in which decline or biomarker status was used as the end point for biomarker discovery, rather than a clinical diagnosis. Doing so, EMIF-AD has developed new treatment targets, multimodality/omics diagnostic tools and qualification level biomarker datasets suitable for presentation to regulatory authorities prior to approval for use in clinical trials and practice.
Finally, prediction rules for cognitive decline in presymptomatic and prodromal AD were developed which will not only improve clinical diagnosis and prognosis, but equally support subject selection and stratification in future clinical trials. These achievements have been possible because EMIF-AD combined both large-scale patient cohorts, linkage with EHR data, and cutting edge biomarker discovery expertise.
Work Packages Overview
Identify predictors of Alzheimer’s Disease (AD) in the pre-clinical and prodromal phase

EMIF-AD Project Achievements
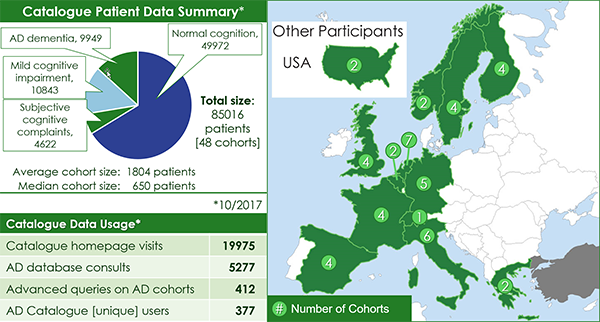
EMIF AD Catalogue
- Tools developed to support data suitability via cohort selection and patient profile selection for research queries
- The user-friendly EMIF Catalogue is now publicly available to search for AD cohorts of interest
- Availability of European AD cohort information in one place will facilitate future research collaboration
- Several initiatives, such as IMI-EPAD, DPUK, IMI-PRISM, JPND-EADB and INTERDEM have already taken up the EMIF Catalogue
The EMIF-AD Multimodal Biomarker Discovery study
Project lead:
- Pieter Jelle Visser, VUmc Amsterdam & Maastricht University
- Simon Lovestone, Oxford University & Janssen Pharmaceutica
- Johannes Streffer, UCB & University of Antwerpen
Coordinator and data management:
- Isabelle Bos, VUmc Amsterdam & Maastricht University, isabelle.bos@maastrichtuniversity.nl
Aim & design:
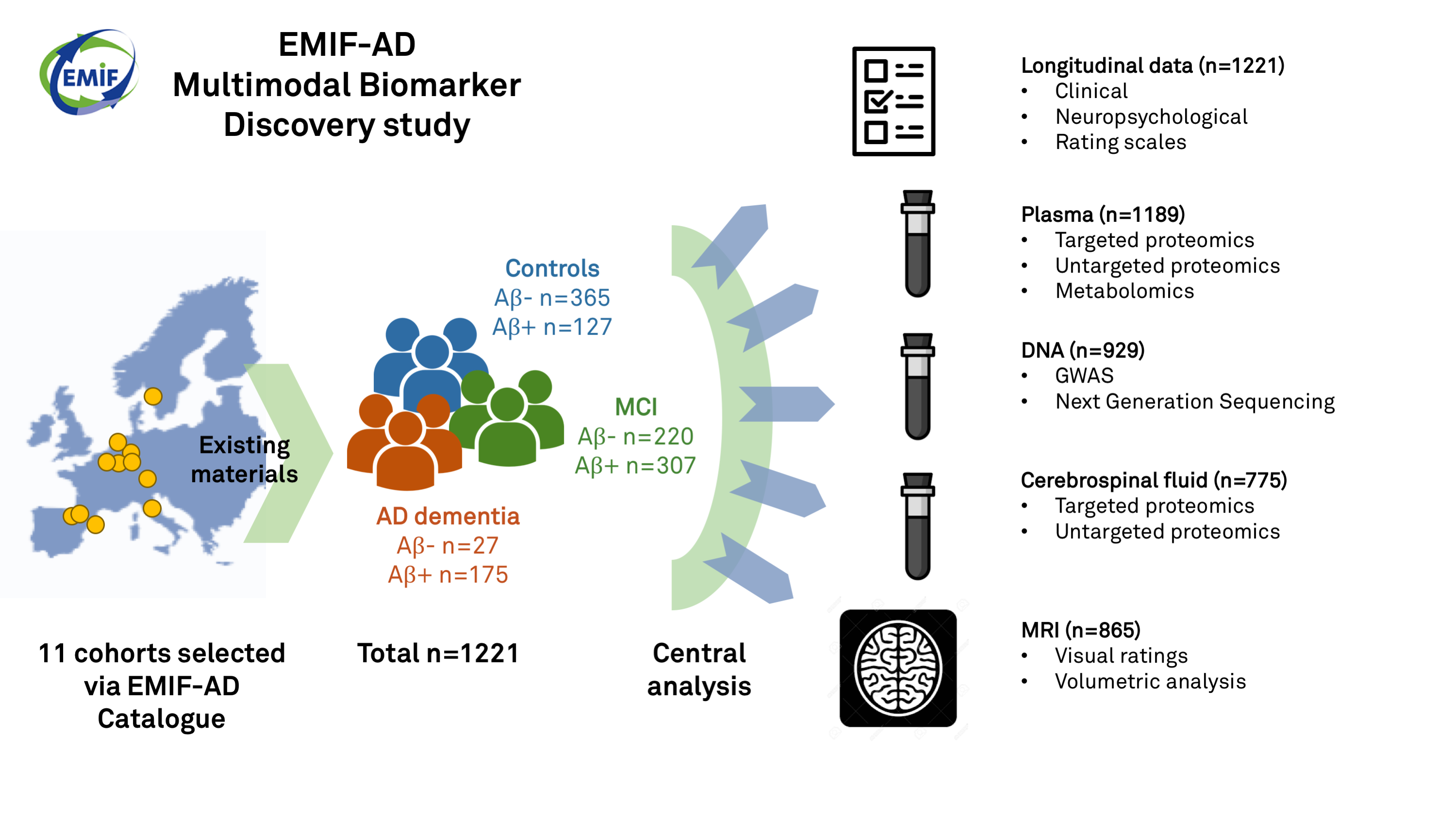
The main aim of the EMIF-AD Multimodal Biomarker Discovery (EMIF-AD MBD) study was to accelerate the discovery of novel diagnostic and prognostic biomarker for AD and to unravel the underlying pathophysiological mechanisms using existing data and samples. We harmonized and pooled clinical data from 11 cohort studies across Europe and samples (cerebrospinal fluid, plasma, DNA) and MRI scans were centrally analyzed using different omics techniques (proteomics, metabolomics, genomics). In total, material from 1221 participants was included (n=492 control, n=527 MCI, n=202 AD dementia). Figure 1 provides a schematic overview of the EMIF-AD MBD study.
The data generated in the context of the EMIF-AD MBD study is available upon request after approval of the research question by all parent cohorts and the EMIF-AD team.
Data requests can be submitted via the EMIF-AD Catalogue (https://emif-catalogue.eu).
Scientific output
- Bos, I., Vos, S.J., Vandenberghe, R., Scheltens, P, Engelborghs, S., Frisoni, G., … Visser, PJ (2018). The EMIF-AD Multimodal Biomarker Discovery Study: Design, methods and cohort characteristics. Alzheimer Research & Therapy. Link
- Kate ten, M., Redolfi, A., Bos, I., Vos, S.J.B., Vandenberghe, R., Gabel, S., … Barkhof, F. (2018) MRI predictors of amyloid pathology: results from the EMIF-AD Multimodal Biomarker Discovery study. Alzheimer Research & Therapy. Link
- Morgan, A. R., Touchard, S., Leckey, C., O’Hagan, C., Nevado-Holgado, A. J., Barkhof, F., … & Engelborghs, S. (2019). Inflammatory biomarkers in Alzheimer’s disease plasma. Alzheimer’s & dementia. Link
- Kim, M., Snowden, S., Suvitaival, T., Ali, A., Merkler, D. J., Ahmad, T., … & Hye, A. (2019). Primary fatty amides in plasma associated with brain amyloid burden, hippocampal volume, and memory in the European Medical Information Framework for Alzheimer’s Disease biomarker discovery cohort. Alzheimer’s & dementia, 15(6), 817-827. Link
- Bos, I., Vos, S., Verhey, F., Scheltens, P., Teunissen, C., Engelborghs, S., … & Bordet, R. (2019). Cerebrospinal fluid biomarkers of neurodegeneration, synaptic integrity, and astroglial activation across the clinical Alzheimer’s disease spectrum. Alzheimer’s & dementia, 15(5), 644-654. Link
The EMIF-AD 90+ study
Project lead:
- Pieter Jelle Visser, VUmc Amsterdam & Maastricht University
- Stephan Carter, University of Manchester
Coordinator and data management:
- Nienke Legdeur & Renée Haan, VUmc Amsterdam, r.haan2@amsterdamumc.nl
Aim & design:
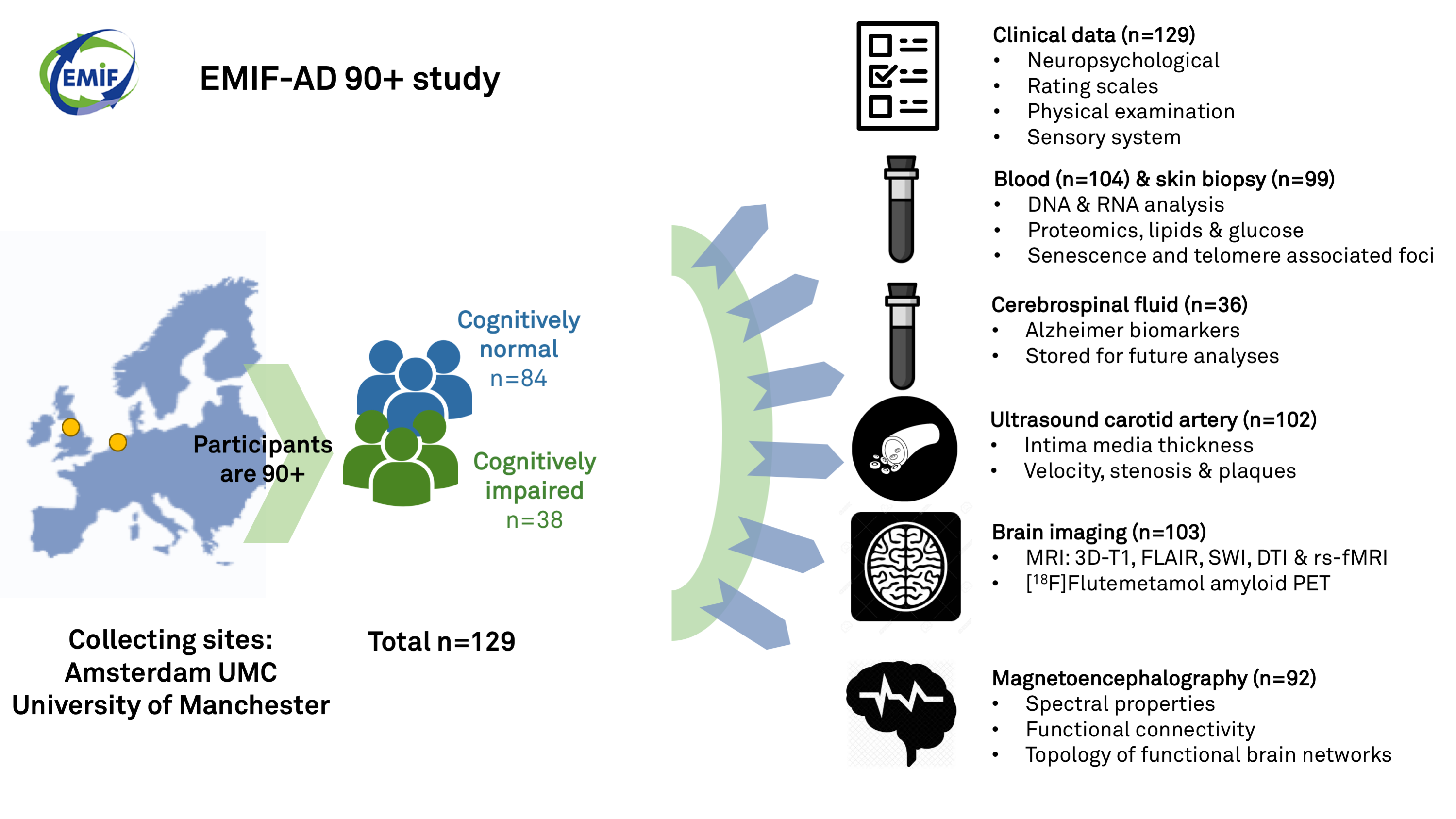
The EMIF-AD 90+ study aimed to identify factors associated with resilience to cognitive impairment in the oldest-old. The study was conducted at the Amsterdam University Medical Center and at the University of Manchester. At baseline, neuropsychological and clinical data (vascular comorbidities, mood, sleep, physical performance, genetic factors) were collected from 129 participants (n=84 with normal cognition and n=38 with cognitive deficits). Regarding imaging measures: baseline MRI (n=92), Amyloid-PET (n=103) and MEG (n=92) have been collected. And in addition skin biopsies (n=99) and ultrasound of the carotid artery (n=102) Currently (Oct 2019), the first annual follow-up measurements have completed in n=129 as of November 2019, with plans for a second annual follow-up 2020.
Data requests can be submitted via the EMIF-AD Catalogue (https://emif-catalogue.eu).
Scientific output
- Legdeur N., Tijms B.M., Konijnenberg E., den Braber A., ten Kate M., Sudre C.H.,…& Visser P.J., Associations of Brain Pathology Cognitive and Physical Markers With Age in Cognitively Normal Individuals Aged 60–102 Years, The Journals of Gerontology: Series A. Link
- Legdeur N., Badissi M., Carter S.F., de Crom S., van de Kreeke A., Vreeswijk R., … & Visser P.J. Resilience to cognitive impairment in the oldest-old: design of the EMIF-AD 90+ study. BMC Geriatr. Link
The EMIF PreclinAD study
Project lead:
- Pieter Jelle Visser, VUmc Amsterdam & Maastricht University
- Stephan Carter, University of Manchester
Coordinator and data management:
- Jori Tomassen, VUmc Amsterdam, j.tomassen@amsterdamumc.nl
Aim & design:
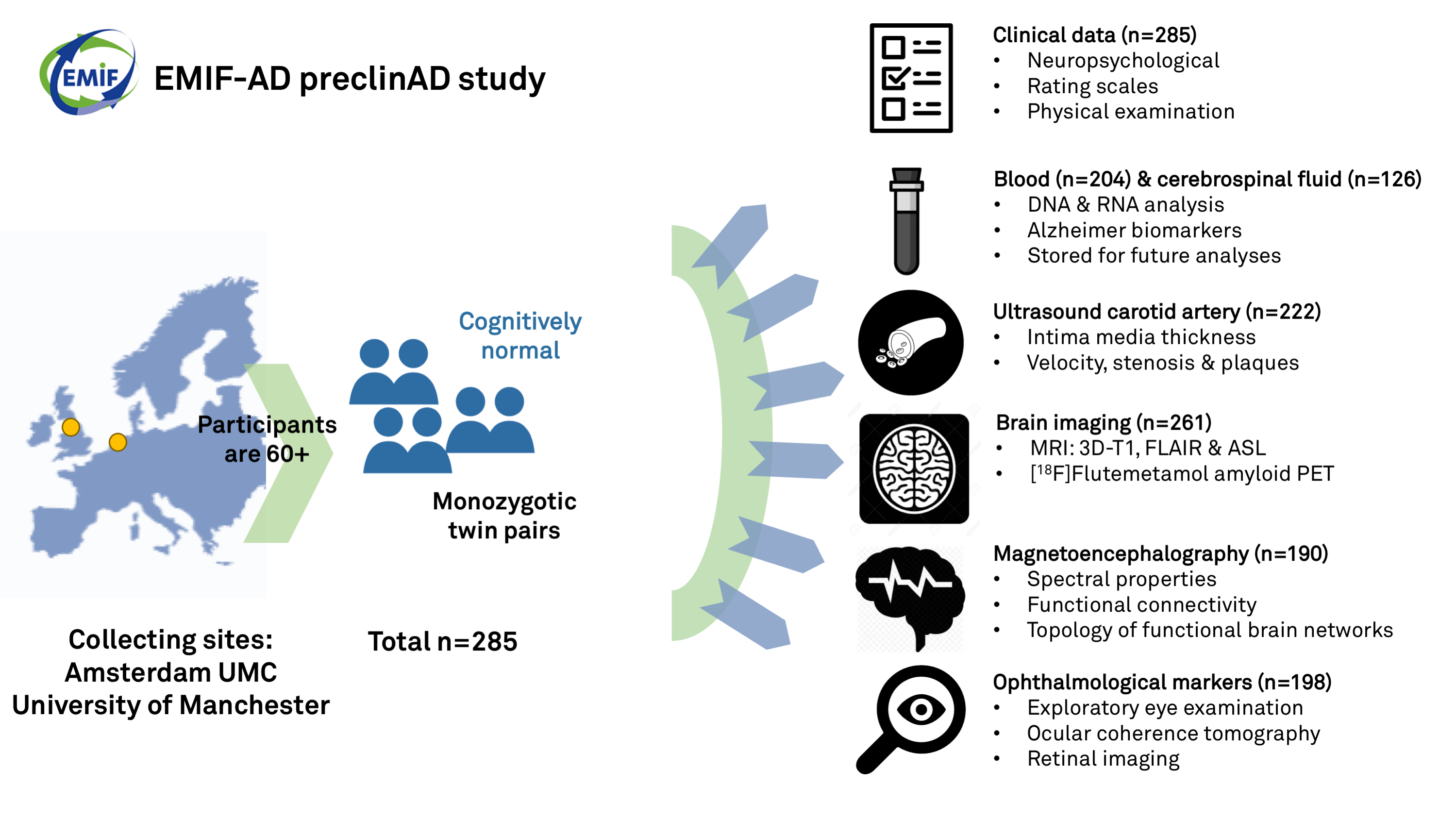
The main aim of the EMIF-AD PreclinAD study was to indentify new risk factors and diagnostic markers for both amyloid pathology and cognitive decline in cognitively normal subjects with or without amyloid pathology. To investigate this monozygotic twin pairs were included such that genetic and environmental pathways can be identified. For the baseline measurement n=204 cognitively healthy elderly monozygotic twins aged 60 years and older were included from the Manchester and Newcastle Age and Cognitive Performance Research Cohort and the Netherlands Twin Register. At baseline the following measurements were done: neuropsychological examination (n=204), blood sampling (n=204), CSF collection (n=127), ultrasound of the carotid artery (n=102), magnetoencephalography (n=190) and collection of ophthalmological markers (n=198). In n=192 (94%) the following follow-up measures were done after two years: containing a neuropsychological examination (n=191), a blood collection (n=192) and a CSF sampling (n=103). A second follow-up will be conducted in 2020.
Data requests can be submitted via the EMIF-AD Catalogue (https://emif-catalogue.eu).
Scientific output
- Konijnenberg E., Carter S.F., Ten Kate M., den Braber A., Tomassen J., Amadi C., … & Visser P.J. The EMIF-AD PreclinAD study: study design and baseline cohort overview. Alzheimers Res Ther. 2018 Aug 4;10(1):75. doi: 10.1186/s13195-018-0406-7. Link
- Konijnenberg E., den Braber A., ten Kate M., Tomassen J., Mulder S.D., Yaqub M., …, & Visser P.J. Association of amyloid pathology with memory performance and cognitive complaints in cognitively normal older adults: a monozygotic twin study. Neurobiology of Aging. 2019 May; 77: 58-65. Link